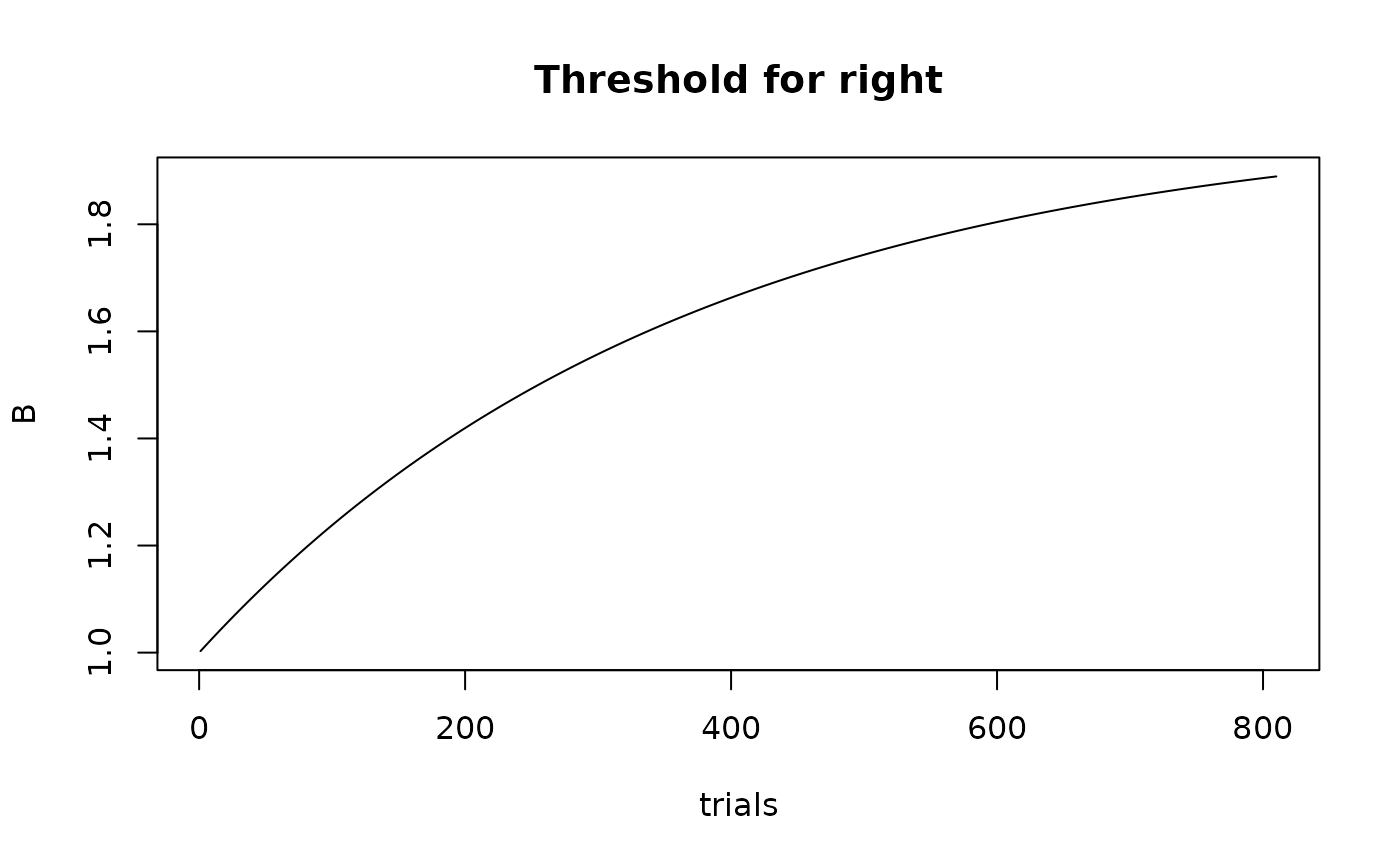
Plots the trend for selected parameters of a model. Can be used either with a p_vector, or trial-wise parameters or covariates obtained from predict()
Usage
plot_trend(
input_data,
emc,
par_name,
subject = 1,
filter = NULL,
on_x_axis = "trials",
pp_shaded = TRUE,
...
)Arguments
- input_data
a p_vector or posterior predictives compatible with the provided emc object
- emc
An emc object
- par_name
Parameter name (or covariate name) to plot
- subject
Subject number to plot
- filter
Optional function that takes a data frame and returns a logical vector indicating which rows to include in the plot
- on_x_axis
Column name in the
dadmto plot on the x-axis. By default 'trials'.- pp_shaded
Boolean. If
TRUEwill plot 95% credible interval as a shaded area. Otherwise plots separate lines for each iteration of the posterior predictives. Only applicable ifinput_dataare posterior predictives.- ...
Optional arguments that can be passed to
plots.
Examples
dat <- EMC2:::add_trials(forstmann)
dat$trials2 <- dat$trials/1000
lin_trend <- make_trend(cov_names='trials2',
kernels = 'exp_incr',
par_names='B',
bases='lin',
phase = "premap")
design_RDM_lin_B <- design(model=RDM,
data=dat,
covariates='trials2', # specify relevant covariate columns
matchfun=function(d) d$S==d$lR,
transform=list(func=c('B'='identity')),
formula=list(B ~ 1, v ~ lM, t0 ~ 1),
trend=lin_trend) # add trend
#> Intercept formula added for trend_pars: B.w, B.d_ei
#> Parameter(s) A, s not specified in formula and assumed constant.
#>
#> Sampled Parameters:
#> [1] "B" "v" "v_lMTRUE" "t0" "B.w" "B.d_ei"
#>
#> Design Matrices:
#> $B
#> B
#> 1
#>
#> $v
#> lM v v_lMTRUE
#> TRUE 1 1
#> FALSE 1 0
#>
#> $t0
#> t0
#> 1
#>
#> $B.w
#> B.w
#> 1
#>
#> $B.d_ei
#> B.d_ei
#> 1
#>
#> $A
#> A
#> 1
#>
#> $s
#> s
#> 1
#>
emc <- make_emc(dat, design=design_RDM_lin_B, compress = FALSE)
#> Processing data set 1
p_vector <- c('B'=1, 'v'=1, 'v_lMTRUE'=1, 't0'=0.1, 'B.w'=1, 'B.d_ei'=1)
# Visualize trend
plot_trend(p_vector, emc=emc,
par_name='B', subject='as1t',
filter=function(d) d$lR=='right', main='Threshold for right')