Plots panels that contain a set of densities for each level of the specified defective factor in the data. These densities are defective; their areas are relative to the respective proportions of the defective factor levels. Across all levels, the area sums to 1. Optionally, posterior/prior predictive densities can be overlaid.
Usage
plot_density(
input,
post_predict = NULL,
prior_predict = NULL,
subject = NULL,
quants = c(0.025, 0.975),
functions = NULL,
factors = NULL,
defective_factor = "R",
n_cores = 1,
n_post = 50,
layout = NA,
to_plot = c("data", "posterior", "prior")[1:2],
use_lim = c("data", "posterior", "prior")[1:2],
legendpos = c("topright", "top"),
posterior_args = list(),
prior_args = list(),
...
)Arguments
- input
Either an
emcobject or a data frame, or a list of such objects.- post_predict
Optional posterior predictive data (matching columns) or list thereof.
- prior_predict
Optional prior predictive data (matching columns) or list thereof.
- subject
Subset the data to a single subject (by index or name).
- quants
Numeric vector of credible interval bounds (e.g.
c(0.025, 0.975)).- functions
A function (or list of functions) that create new columns in the datasets or predictives
- factors
Character vector of factor names to aggregate over; defaults to plotting full data set ungrouped by factors if
NULL.- defective_factor
Name of the factor used for the defective CDF (default "R").
- n_cores
Number of CPU cores to use if generating predictives from an
emcobject.- n_post
Number of posterior draws to simulate if needed for predictives.
- layout
Numeric vector used in
par(mfrow=...); useNAfor auto-layout.- to_plot
Character vector: any of
"data","posterior","prior".- use_lim
Character vector controlling which source(s) define
xlim.- legendpos
Character vector controlling the positions of the legends
- posterior_args
Optional list of graphical parameters for posterior lines/ribbons.
- prior_args
Optional list of graphical parameters for prior lines/ribbons.
- ...
Other graphical parameters for the real data lines.
Examples
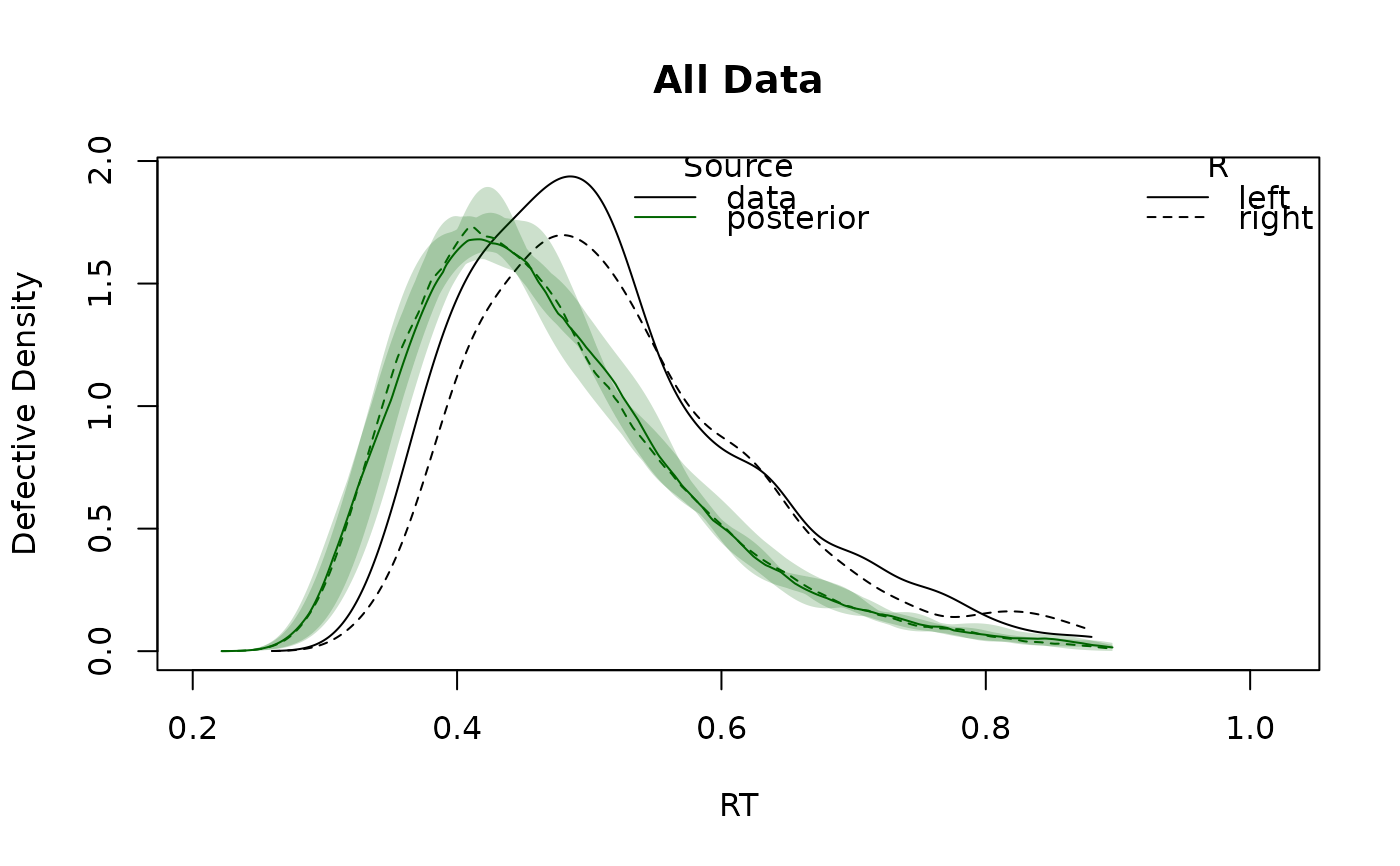
# Plot defective densities for each subject and the factor combination in the design:
plot_density(forstmann)
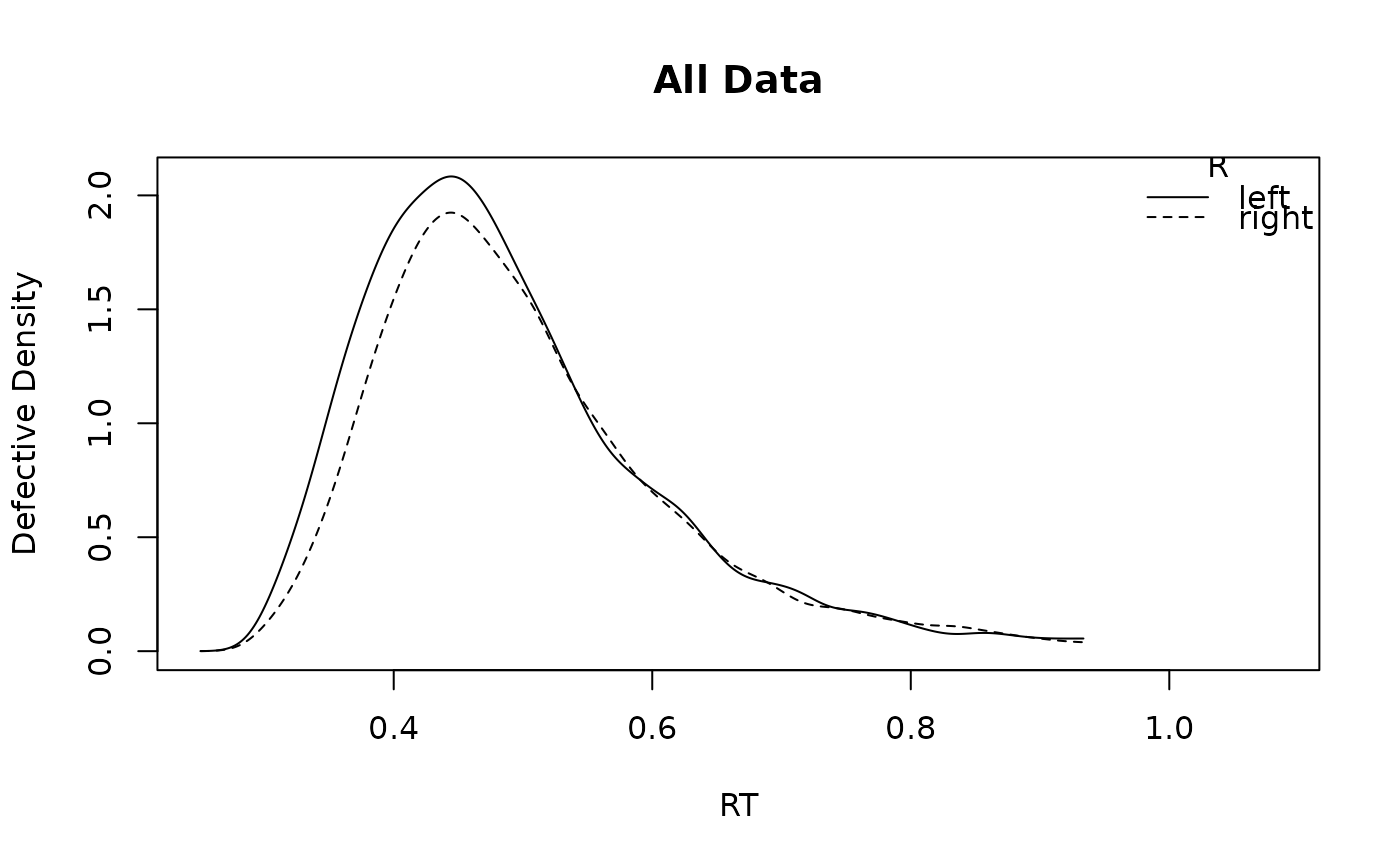 # or for one subject:
plot_density(forstmann, subject = 1)
# or for one subject:
plot_density(forstmann, subject = 1)
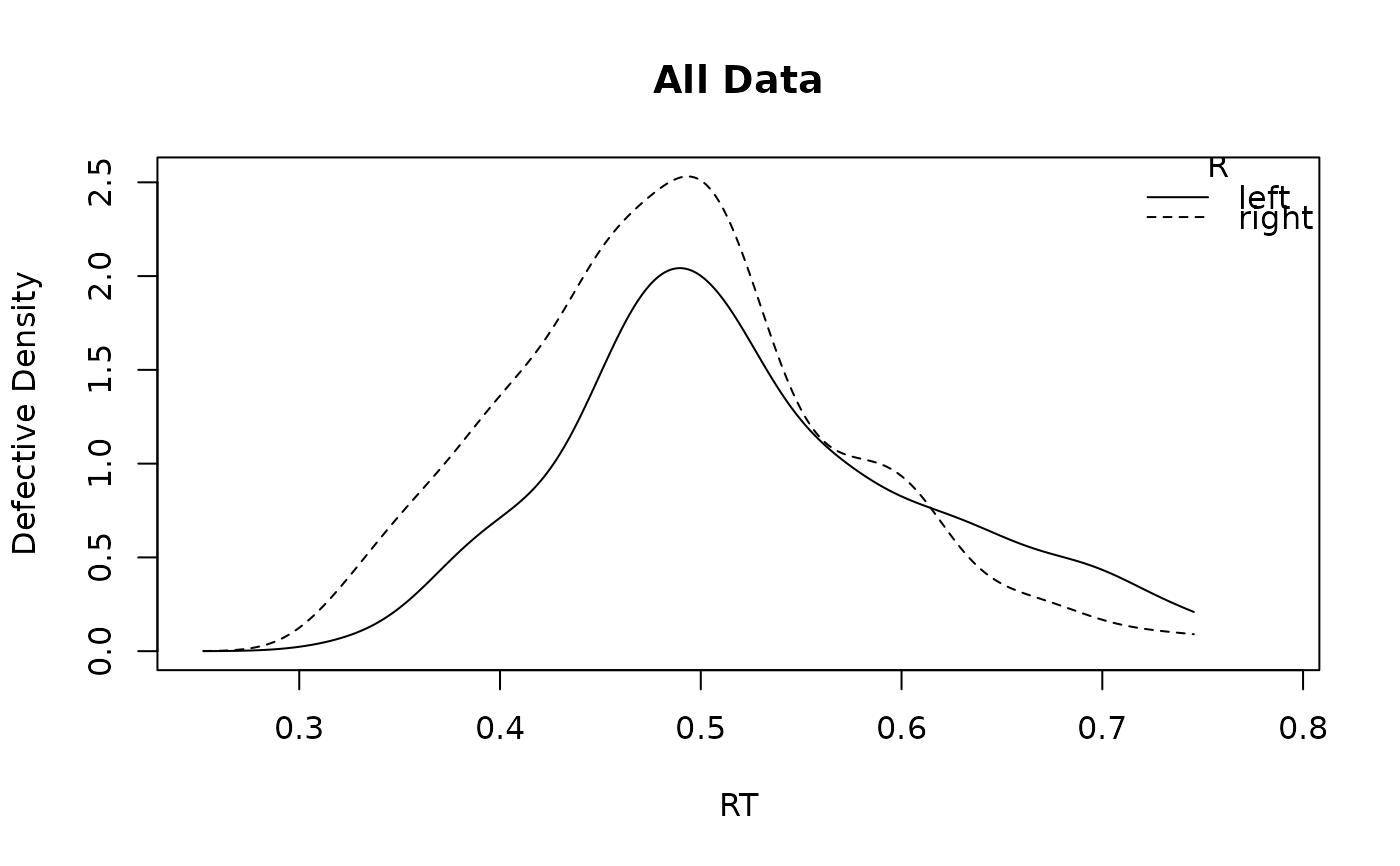 # Now collapsing across subjects and using a different defective factor:
plot_density(forstmann, factors = "S", defective_factor = "E")
# Now collapsing across subjects and using a different defective factor:
plot_density(forstmann, factors = "S", defective_factor = "E")
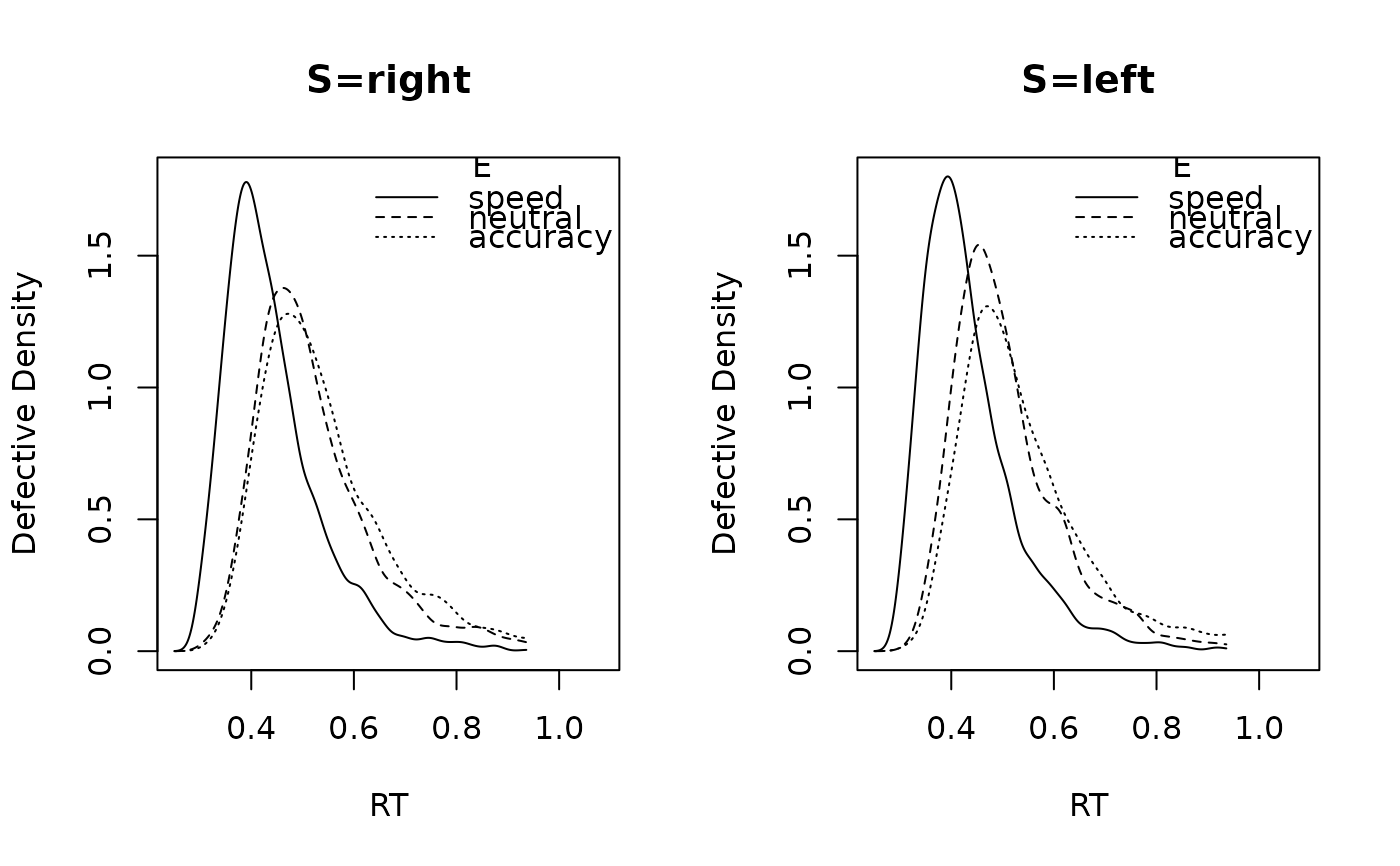 # Or plot posterior predictives
plot_density(samples_LNR, n_post = 10)
# Or plot posterior predictives
plot_density(samples_LNR, n_post = 10)