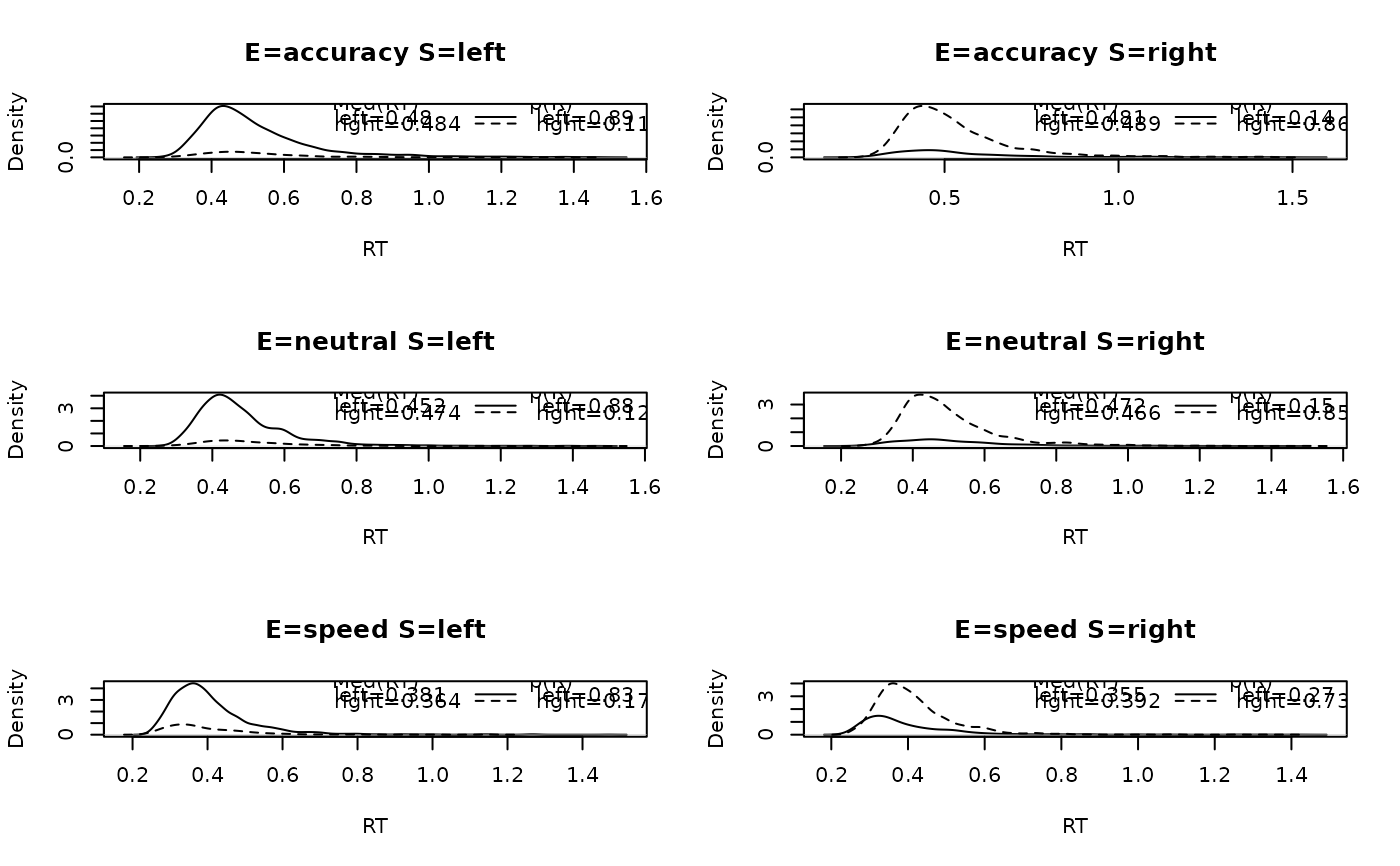
Plots panels that contain a set of densities for each response option in the data. These densities are defective; their areas are relative to the respective response proportion. Across all responses, the area sums to 1.
Usage
plot_defective_density(
data,
subject = NULL,
factors = NULL,
layout = NA,
correct_fun = NULL,
rt_pos = "top",
accuracy = "topright",
...
)Arguments
- data
A data frame. The experimental data in EMC2 format with at least
subject(i.e., the subject factor),R(i.e., the response factor) andrt(i.e., response time) variable. Additional factor variables of the design are optional.- subject
An integer or character string selecting a subject from the data. If specified, only that subject is plotted. Per default (i.e.,
NULL), all subjects are plotted.- factors
A character vector of the factor names in the design to aggregate across Defaults to all (i.e.,
NULL).- layout
A vector indicating which layout to use as in par(mfrow = layout). If NA, will automatically generate an appropriate layout.
- correct_fun
If specified, the accuracy for each subject is calculated, using the supplied function and an accuracy vector for each subject is returned invisibly.
- rt_pos
legend function position character string for the mean response time (defaults to
top)- accuracy
legend function position character string for accuracy (defaults to
topright)- ...
Optional arguments that can be passed to
get_pars,density, orplot.default(seepar())
Examples
# First for each subject and the factor combination in the design:
plot_defective_density(forstmann)
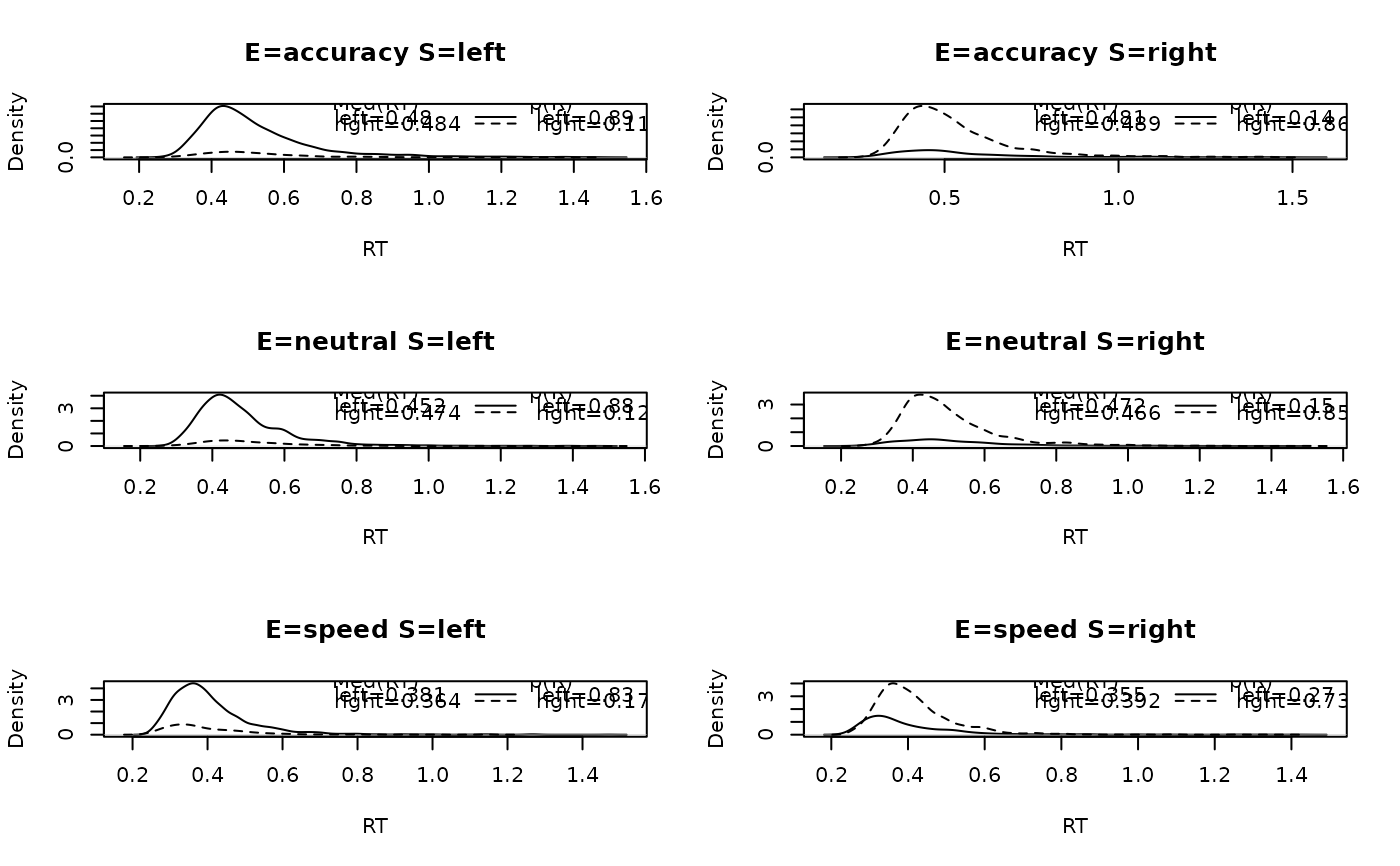 # Now collapsing across subjects:
plot_defective_density(forstmann, factors = c("S", "E"))
# Now collapsing across subjects:
plot_defective_density(forstmann, factors = c("S", "E"))
 # If the data is response coded, it generally makes sense to include the "S" factor
# because EMC2 will plot the "R" factor automatically. This way, choice accuracy can
# be examined
# Each subject's accuracy can be returned using a custom function:
print(plot_defective_density(forstmann, correct_fun = function(d) d$R == d$S))
# If the data is response coded, it generally makes sense to include the "S" factor
# because EMC2 will plot the "R" factor automatically. This way, choice accuracy can
# be examined
# Each subject's accuracy can be returned using a custom function:
print(plot_defective_density(forstmann, correct_fun = function(d) d$R == d$S))
 #> as1t bd6t bl1t hsft hsgt kd6t kd9t kh6t
#> 0.7666667 0.8386337 0.7947805 0.9375000 0.9552415 0.6855124 0.9623086 0.8661888
#> kmat ku4t na1t rmbt rt2t rt3t rt5t scat
#> 0.8947991 0.7207101 0.9551887 0.8652226 0.9180523 0.7520759 0.6483357 0.7926973
#> ta5t vf1t zk1t
#> 0.9195266 0.8436754 0.8114144
#> as1t bd6t bl1t hsft hsgt kd6t kd9t kh6t
#> 0.7666667 0.8386337 0.7947805 0.9375000 0.9552415 0.6855124 0.9623086 0.8661888
#> kmat ku4t na1t rmbt rt2t rt3t rt5t scat
#> 0.8947991 0.7207101 0.9551887 0.8652226 0.9180523 0.7520759 0.6483357 0.7926973
#> ta5t vf1t zk1t
#> 0.9195266 0.8436754 0.8114144